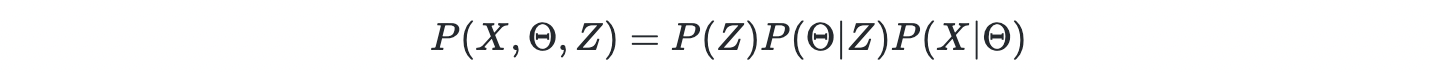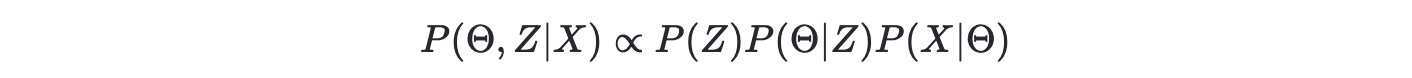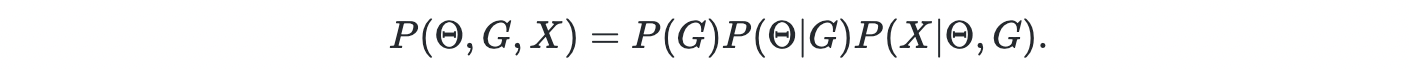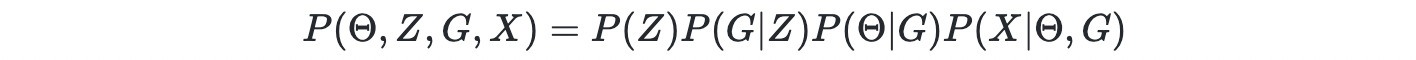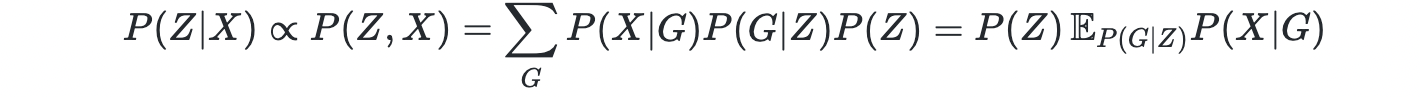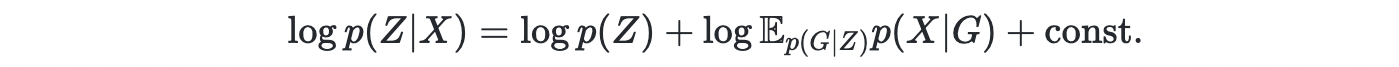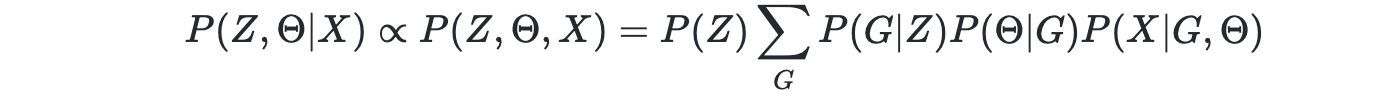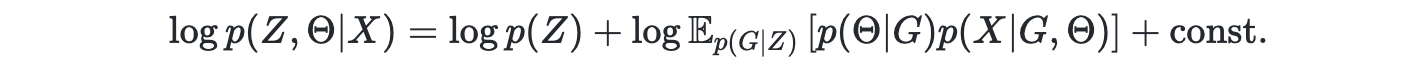Massive projects in multi-modal learning, self-supervised learning, and 3D imagery were definitely the stars of the recent NeurIPS 2021. However, one brain candy titled "Differentiable Bayesian Structure Learning" in causal inference session caught my attention. Causal inference has always been a fascinating topic to me. Why did the stock markets fluctuate so dramatically today? Which stocks were causing the stock movements of other stocks? What are the gene regulation mechanisms in different cancer cells? What are the toxic metabolism pathways that give rise to diabetes? What are the signaling pathways that human employs to combat virus infections? If you are also intrigued by such questions, then you got a scientific mindset and should love to learn more about causal inference. Yet, the causal inference formalism is mathematically intimidating and not scalable to large datasets. Fortunately, authors of this paper presented a scalable and differentiable approach to this issue and they synthesized multiple pieces of elegant mathematics into one powerful algorithm. In addition, one of my favorite machine learning scientists and godfather of kernel method, Bernhard Schölkopf, was also among the authors of this awesome work. Now allow me to walk you through the key ideas of this work.
TL;DR
- Bayesian structure learning
- Bayesian modulo frequentist: distribution of model parameter
- Bayesian network (BN)
- discrete dependency graph, i.e. directed acyclic graph (DAG)
- continuous parameters
- Graph
- node embedding is similar to word embedding
- but asymmetric with respect to in and out message
- a differentiable constrain of acyclic graph, or DAGness penalizer
- graph priors: edge or degree distribution
- Variational inference
- SVGD: Stein variational gradient descent
- or optionally traditional variational inference
Introduction
Deep learning has already made great impacts in a wide range of engineering fields, such as image processing, video processing, text understanding, speech recognition and synthesis, and etc. In recent years, deep learning has also been reshaping the landscape of scientific research. The heart of science is discovering causal relations between subjects of interest. This paper discussed a method that automatically captured the causal relations within a protein signaling network using Bayesian network and a novel type of gradient descent. The models are not trillion-parameter monsters but they are able to capture important relations within a network at scale and could potentially revolutionize biology and medical research.
The goal of Bayesian structure learning is learning a full posterior distribution of Bayesian networks (BN) from the observations where one of the key challenge is working with a joint distribution over the space of discrete directed acyclic graphs and continuous conditional distribution parameters.
A Bayesian network (also known as a Bayes network, Bayes net, belief network, or decision network) is a probabilistic graphical model that represents a set of variables and their conditional dependencies via a directed acyclic graph (DAG).
DAG
A graph consists a set of nodes and a set of edges that connect the nodes. The edges of a directed graph are asymmetric "arrows" instead of symmetric "bonds." The idea of being acyclic can be interpreted by random walk over the DAG. Let's say we put a drunkard at any one of the DAG node and allow him to randomly pick directions as long as there is an arrow connecting current node and next node. If for whatever starting node, however he tries, he is not able to return to the original node, then the directed graph is a bona fide DAG. The above statement can be mathematically expressed using matrix exponential of adjacency matrix G whose trace is equal to number of nodes.
Bayesian inference in a nutshell
In Bayesian perspective, everything is a random variable including the observations X, model parameters Θ, and latent states Z. Let's say the latent states determines the model parameters Z→Θ, and the model parameters predict the changes of observation Θ→X. We can start with joint density of all the random variables,
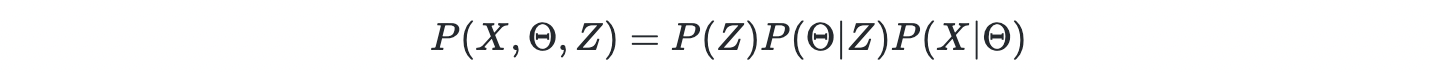
where P(X∣Θ) is known as the likelihood or chance of observing X given model parameter Θ; P(Θ∣Z) is a prior of the likelihood; P(Z) is the prior for the P(Θ∣Z). The posterior is the degree of belief of the "invisible" random variables given observations
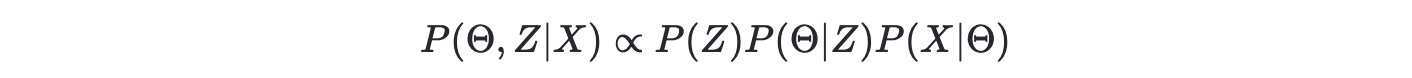
up to a constant denominator of total evidence.
Connect the dots
A Bayesian network is graphical model where the "graph" is a DAG where the nodes represent random variables and arrows indicate causal relations between the random variables. The parameters of a BN include the DAG G and model parameters Θ. The joint density can be expanded as follows
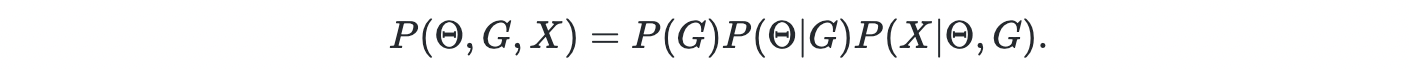
However, the space of DAG is discrete and exponentially large. Thus, it is challenging to learn the DAG using gradient descent. Thus, the authors introduced a continuous latent variable Z i.e. graph embedding, as the parent variable of graph G. Now the joint density becomes
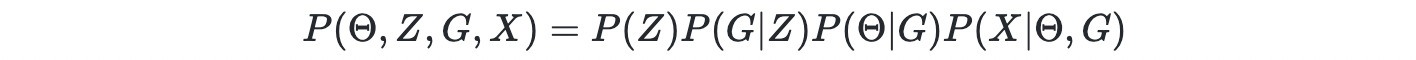
For a simple case where P(X∣G) can be computed directly, the posterior can be computed using
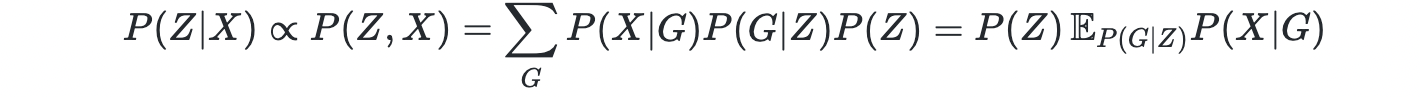
and
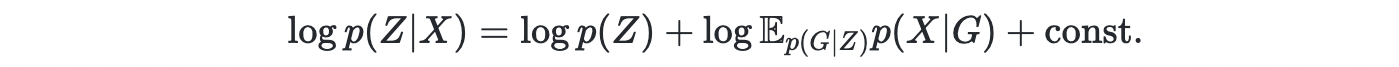
In general, Z and Θ are learned jointly,
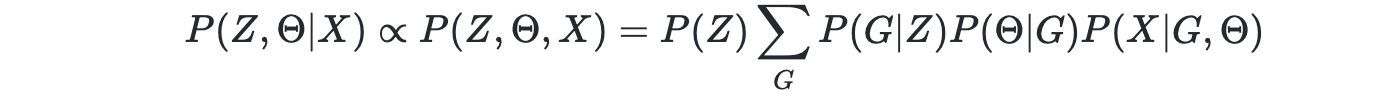
and
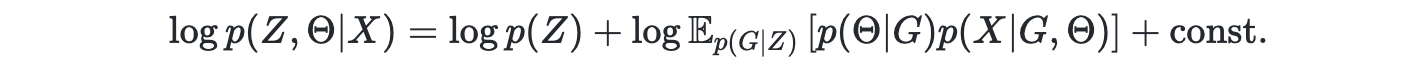
Stein Variational Inference
In order to learn the latent variable Z and model parameter Θ using gradient descent, one need to minimize the KL-divergence between the posterior and a tractable ansatz distribution. Instead of taking the parametric approach e.g. the mean-field exponential family ansatz, the authors chose a non-parametric kernel-based ansatz which is very similar to particle filter and kernel density estimators and is able to approximate generic distributions given sufficient number of particles. The algorithm is known as Stein gradient descent. Its awesomeness deserves another blog post to elaborate.
Conclusions
In summary, the authors introduced a novel approach to learn Bayesian networks using gradient descent. One of the most important contribution is that the combinatorially large discrete graph space was replaced by a continuous latent space with a differentiable soft DAGness penalty. Another highlight of this work is the non-parametric Stein variational inference which allows for multi-modal distributions of latent graph embedding.
References
[1] DiBS: Differentiable Bayesian Structure Learning
[2] DAG-GNN: DAG Structure Learning with Graph Neural Networks
[3] Stein Variational Gradient Descent: A General Purpose Bayesian Inference Algorithm
[4] Stein Variational Gradient Descent as Moment Matching